
Perez Lab
Mechanistic biology of cell wall assembly and membrane proteins
Welcome to the Perez Lab
The bacterial cell wall is a complex matrix assembled through the activity of multiple pathways involving membrane proteins. Understanding their molecular mechanism is essential to unravel the mysteries behind this important defense barrier. My laboratory aims to elucidate the mechanism of transporters, flippases and polymerases involved in the assembly of the cell wall, reveal how their function influences bacterial adaptation, and redirect the activity of these proteins towards applications in the synthesis of glycoconjugates. Read more on our Research page.

Selected Publications
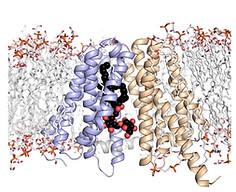
Receptor-recognition and antiviral mechanisms of retrovirus-derived human proteins. Link to publication in NSMB In collaboration with the Reyes Lab we present cryo-electron microscopy structures and functional characterization of human ASCT2 in complexes with the receptor-binding domains of syncytin-1 and suppressyn, cellular proteins of retroviral origin involved in cell–cell fusion events to establish the maternal–fetal interface in the placenta. Our work uncovers a common ASCT2 recognition mechanism by a large group of endogenous and disease-causing retroviruses, and provides high-resolution views on how placental human proteins exert morphological and immunological functions.
Mechanistic basis of choline import involved in teichoic acids and lipopolysaccharide modification
Link to publication in Sci. Adv.
We determined the structure and mechanism of LicB, a choline transporter involved in the cell wall biosynthesis of Streptococcus pneumoniae. We used a multidisciplinary approach including structure determination by cryo-EM and X-ray crystallography, electrophysiology, in-vitro transport assays, sybody generation in collaboration with the Seeger Lab, and in- vivo assays in collaboration with the Veening Lab.
Evidence for a trap-and-flip mechanism in a proton-dependent lipid transporter
Link to publication in Nat. Comm.
We investigated the flipping mechanism of LtaA, a proton-dependent MFS lipid transporter essential for lipoteichoic acid biosynthesis in the pathogen Staphylococcus aureus. Combining cysteine disulfide trapping, mutagenesis, transport assays in vitro and in vivo, and molecular dynamics simulations in collaboration with the Mehdipour Lab and Hummer Lab, we revealed that LtaA catalyzes lipid translocation by a trap-and-flip mechanism.
Structure of a proton-dependent lipid transporter involved in lipoteichoic acids biosynthesis
Link to publication in NSMB
We elucidated the first structure a lipid transporter from the MFS superfamily of transporters. LtaA, a flippase involved in lipoteichoic acid assembly in Staphylococcus aureus, catalyse lipid export by import of protons, which contributes to the adaptation of S. aureus to acidic stress conditions.



Selected Recent and Upcoming Talks
November 2026, Georgia State University, Atlanta, GA
July 2026, The Protein Society Annual Symposium, Boston, MA, USA
March 2026, Gordon Research Conference of Ligand Recognition and Molecular Gating, Lucca, Italy
February 2026, University of Alabama Birmingham, Birmingham, AL
November 2025, Society for Glycobiology, San Diego, CA, USA
Current Funding
-
NIH-NIGMS (Since 2025)
-
NIH-NIDCR (Since 2025)
-
University of Georgia (Since 2024)
-
SNSF project (Since 2022)




